I have a gene list. How do I convert my gene IDs to FoPDGB gene IDs?
How many genomes are included in the Pangenome analysis?
JBrowse is not working, what should I do?
I have two genomes; how can I generate alignments such as the ones under the Align section?
How do I use various functions?
(1) I have a gene list. How do I convert my gene IDs to FoPDGB gene IDs?
There are two ways.
(a) Convert gene IDs to FoPGDB gene IDs.
For genomes deposited to NCBI: The gene IDs in FoPDGB are the same as gene-locustag IDs in NCBI.
For genomes deposited to JGI and NGDC: Gene IDs in FoPDGB consist of genome code and their original gene IDs in gff file, just like ‘Fob72_gene_1’. The ‘Fob72’ is the genome code of Fusarium oxysporum f. sp. basilici Amherst-72 v1.0 and the ‘gene_1’ is the gene IDs in gff files. It's more convenient to use BLAST in (b).
(b) Find gene ID in FoPGDB through BLAST.
If you have a DNA or protein sequence, you can use the BLAST in the tool section to align the sequence with the sequences in the database, which could help you find the gene name of this sequence in our database.
(2) How many genomes are included in the Pangenome analysis?
We collected 35 high-quality genomes of Fusarium oxysporum from NCBI, JGI, and NGDC. You can find the detailed information including Genome code, race, reference, download link, and number of genes and sequences on the “Genome” page.
(3) JBrowse is not working, what should I do?
Sometimes you could meet the problem of not loading SNP&INDEL.vcf and SV.vcf information in JBrowse, which is because the amount of SNP&INDEL.vcf and SV.vcf is too large. The easy way to solve this problem is to control the size of the region you are looking at and not move the browser too fast.
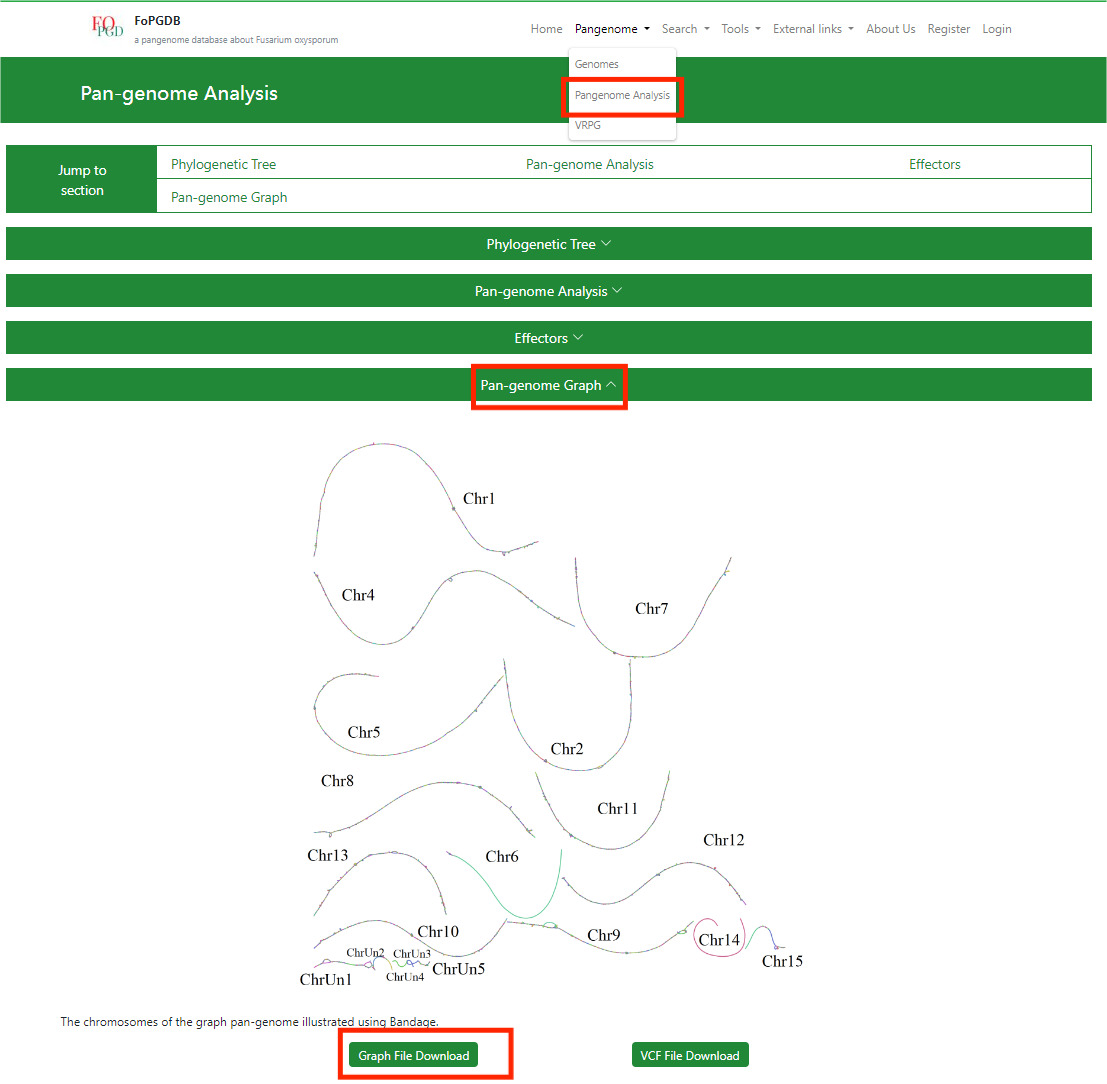
(4) I have two genomes; how can I generate alignments such as the ones under the Align section?
The genome alignments in the Align section are generated using D-Genies you can upload your genome sequence files to generate similar plots.
(5) How do I use various functions?
Here is A case study with the TOR kinase gene.
(a) Fing gene ID using BLAST
Firstly, you should input the sequence into the checkbox or input the sequence file and choose the databases you want to use. If you have other requirements, you can adjust some parameters in the advanced options area.
Then, you can click the "BLAST" button.

Finally, when you finish these steps, you can get the BLAST result, a list of the sequences in the database you choose. These sequences have high similarity with the sequence you input. The sequence on the top of the list has the largest similarity with the input sequence, so this sequence’s name is the name of the input sequence in our database.
You can download the results in Alignment, Tab-delimited, GFF3, and XML formats.

(b) Batch search to check core/dispensable.
You can see the information related to this gene with the “Batch Search” function in the “Search” module. By inputting the gene name into the checkbox and clicking the submit button, you can get the information about this gene in our database.
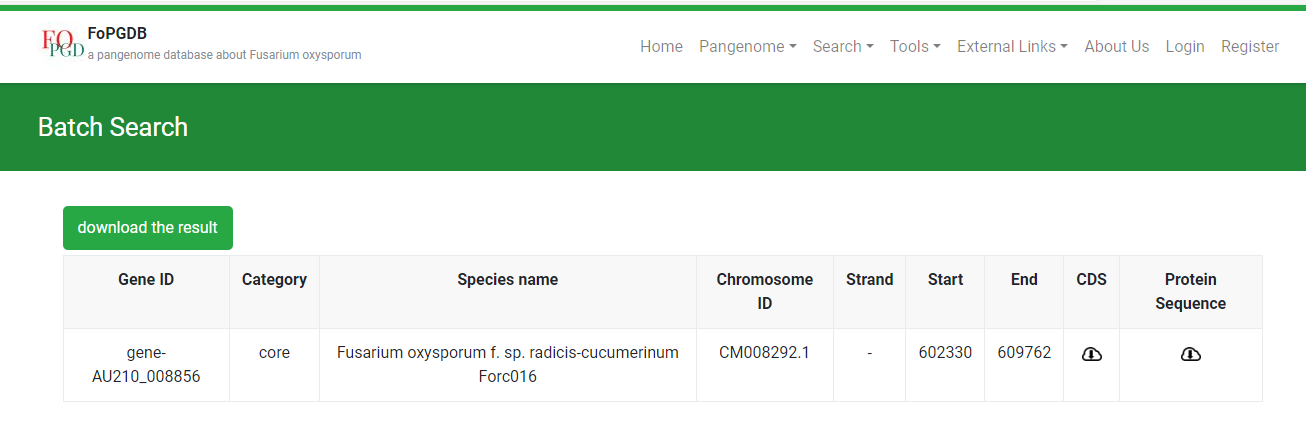
Here is the search result. From the picture below, you can see this gene’s position, category, CDS sequence, and so on.

(c) Effector check
You can also check some effector prediction results’ information about this gene with the effector search function in the search section. By inputting the gene’s name into this checkbox, we can get the effector prediction results of some popular software, such as SignalP, TargetP, and EffectorP.

(d) Check orthologous genes
We can also see other genes in the same group with this gene with the Orthogroup Search function in the search section. Firstly, you should input the gene name into the input box on the left side, and then click the submit button. You can get the search results.

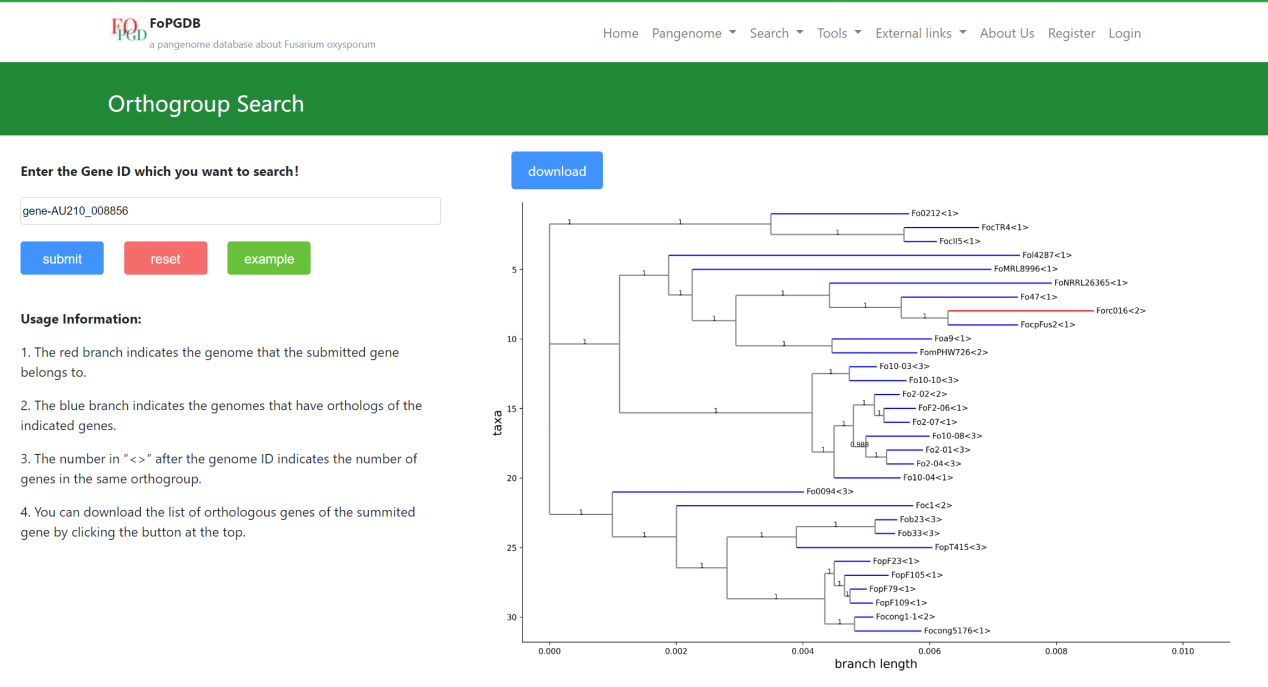
In the search result, you can see the distribution of the same orthogroup’s genes in the phylogenetic tree. The red branch indicates the genome that the submitted gene belongs to and the blue branch indicates the genomes that have orthologs of the indicated genes. The number in the <> means the number of this orthogroup’s genes in this genome. Besides, you can also download the CSV file about details of these genes using the download button.
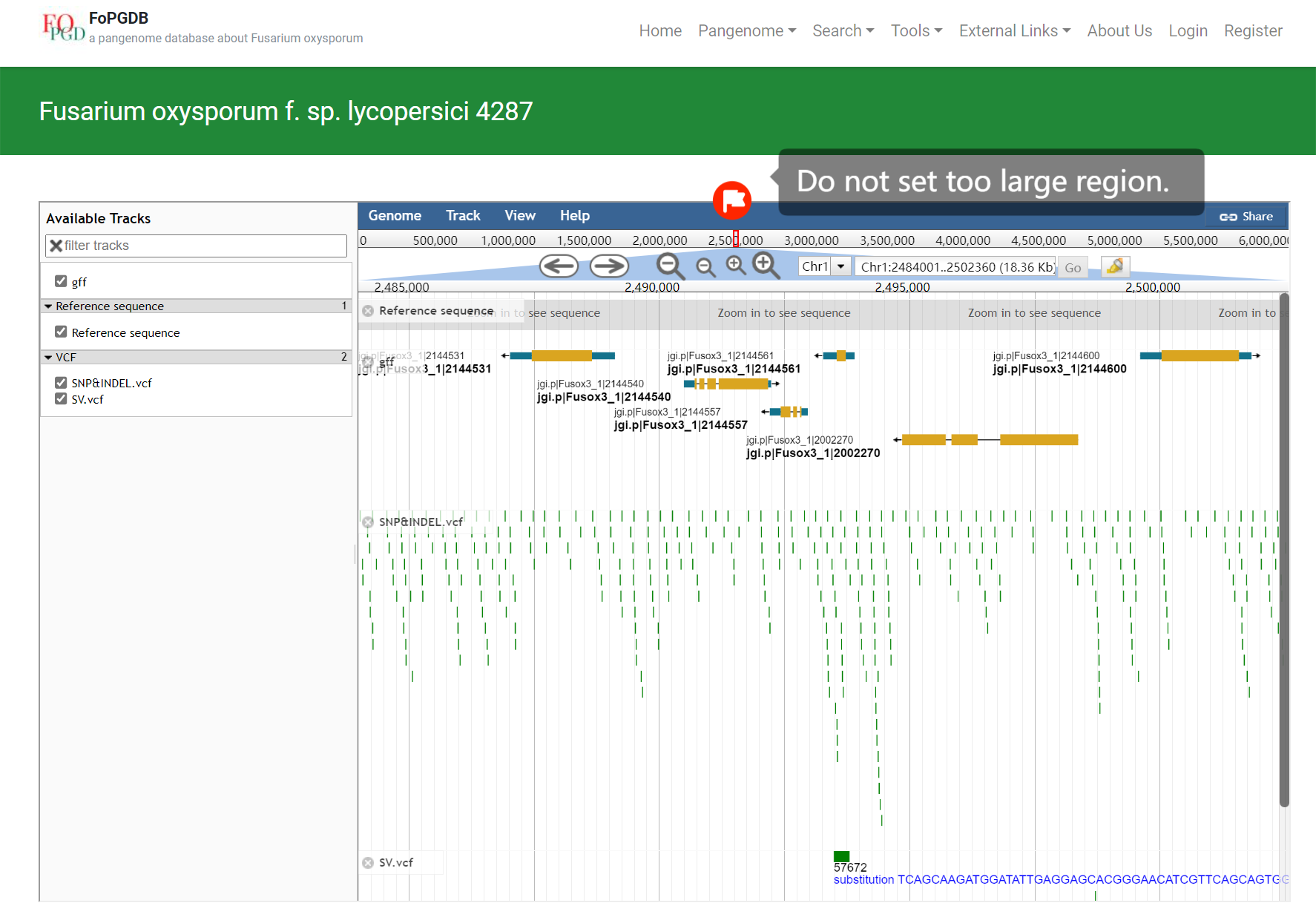
(e) Browse genome locus via JBrowse
You can also see the gene sequence’s details with the function JBrowse in the Tools section. Here are the steps.
e.1 With the Batch Search function, you have got the position of this gene. Choose the genome to which the gene belongs and click it.

e.2 Then choose the chromosome and region of this gene.

e.3 Finally, when the region is limited to a small region, you can click the options of SNP&INDEL.vcf and SV.vcf. Sequence, gene model, SVs, SNPs, INDELs, and transposable elements are shown in the JBrowse.

(f) Check the graph
Owing to the pan-genome graph constructed based on the Fol4287, you need to find the gene in Fol4287 which belongs to the same orthogroup with the TOR kinase gene. Therefore you need to check the result of the orthogroup search function and you can find the Fol4287_gene_10062 is the gene you need.
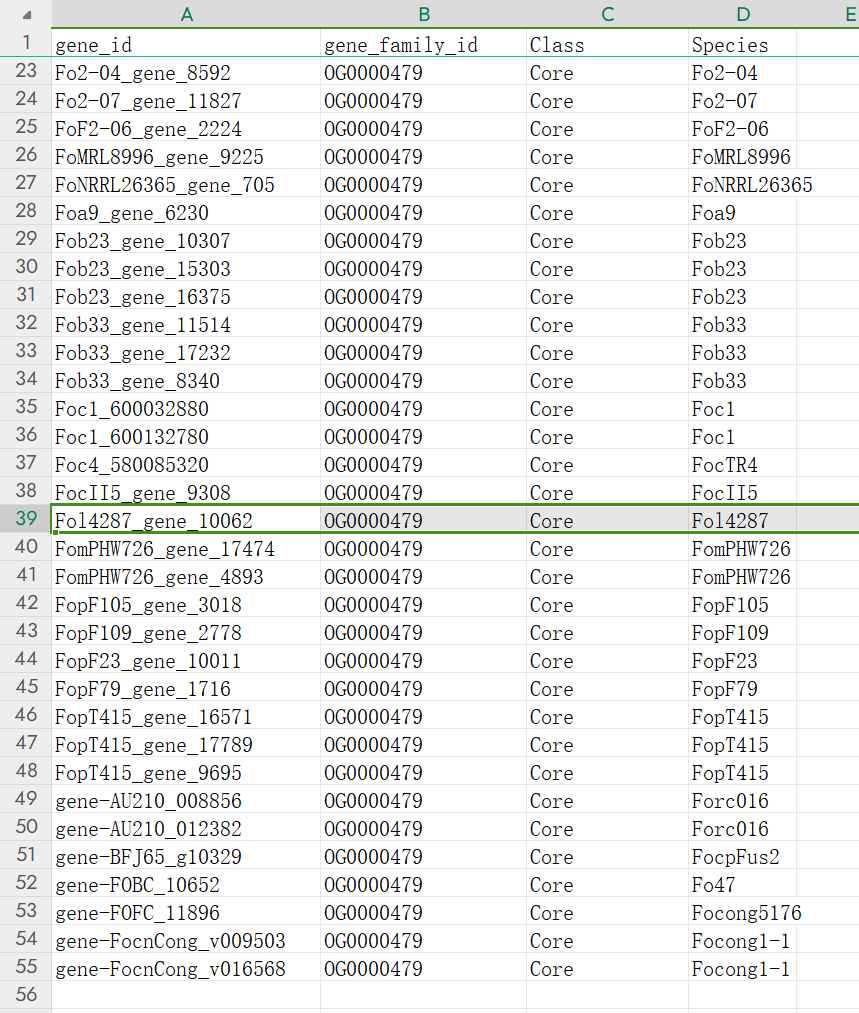
Then you need to use the batch search to find the gene’s position.


Finally, you can use the VRPG function in the Pangenome section to see the gene in the pangenome graph. You should choose the chromosome, starting point, and end point. Then you can see the result. Here, we observe some structural variations near the gene however the region where the gene is located is conserved in the pangenome.

(g) Finally, you can download the pangenome graph to do various analyses such as genotyping the SNPs, INDELs, and SVs using short-read sequencing data.